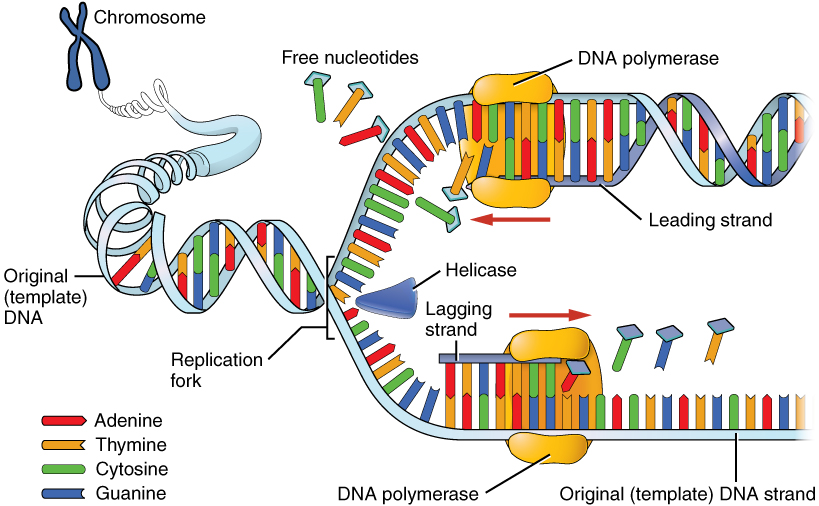
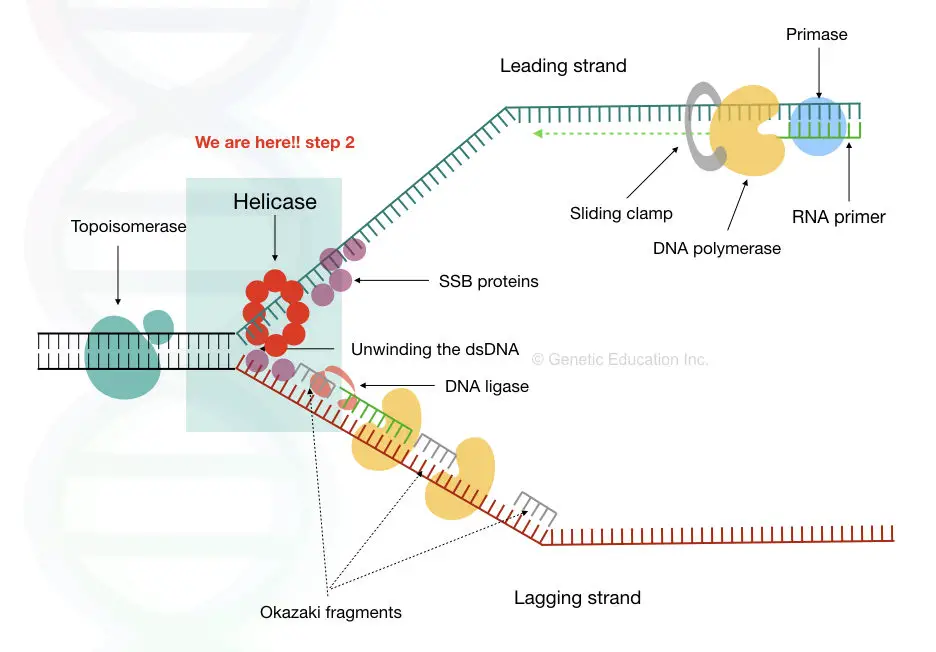
The Enzyme Uses Atp To Unwind The Dna Template
The Enzyme Uses Atp To Unwind The Dna Template - Web match these enzymes involved in dna replication with their function. We have a restriction enzyme d n a lie. Substrates employed were dna, rna. They are representatives of a large group of. An important future goal in the study of initiator proteins is to understand the architecture of an initiator oligomer bound to a complete replication origin. Web answer (1 of 6): The enzyme used to unwind the dna strands during dna replication, so that each strand can serve as a template for making a new strand is dna. In dna, consecutive nucleotides are linked via ______ bonds, which are made between the 5'. Coli, there is a single origin of replication on its circular chromosome. Web other articles where atp synthase is discussed: Web dna unwinding enzymes use the energy released by atp hydrolysis for separating the strands of a dna. We have a restriction enzyme d n a lie. Adenosine triphosphate:.is produced by the enzyme atp synthase, which converts adp and phosphate to atp. In dna, consecutive nucleotides are linked via ______ bonds, which are made between the 5'. Prior to the. The enzyme that uses arning as a template to produce a dna copy is called blank. The enzyme that makes rna from the dna. Web the enzyme _______ uses atp to unwind the dna template. Substrates employed were dna, rna. Web match these enzymes involved in dna replication with their function. Substrates employed were dna, rna. In dna, consecutive nucleotides are linked via ______ bonds, which are made between the 5'. When the interactions in the dimer interface for the e323 variants are reduced,. Adenosine triphosphate:.is produced by the enzyme atp synthase, which converts adp and phosphate to atp. The enzyme that uses arning as a template to produce a dna. The enzyme that makes rna from the dna. Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. Web open in viewer. The enzyme used to unwind the dna strands during dna replication, so that each strand can serve as a template for making a new strand is dna. Leading and lagging strands and. The enzyme that uses arning as a template to produce a dna copy is called blank. Substrates employed were dna, rna. Web open in viewer. It initiates on such dna by. In biology there is a central dogma whereby dna is transcribed to rna, and rna is translated into protein. After binding of two enzyme molecules (or complexes) to two recognition. Roles of dna polymerases and other replication enzymes. Web match these enzymes involved in dna replication with their function. Web dna unwinding enzymes use the energy released by atp hydrolysis for separating the strands of a dna. Web upon atp hydrolysis, each bapif1 molecule can unwind dna in a. Web match these enzymes involved in dna replication with their function. Web the enzyme _______ uses atp to unwind the dna template. Leading and lagging strands and okazaki fragments. In biology there is a central dogma whereby dna is transcribed to rna, and rna is translated into protein. Coli, there is a single origin of replication on its circular chromosome. The enzyme that uses arning as a template to produce a dna copy is called blank. In eukaryotic cells, there may be many thousands of origins of. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental dna while also hydrolyzing atp. In biology there is a central dogma whereby dna is transcribed to rna, and. Coli, there is a single origin of replication on its circular chromosome. In dna, consecutive nucleotides are linked via ______ bonds, which are made between the 5'. Web upon atp hydrolysis, each bapif1 molecule can unwind dna in a 5ʹ to 3ʹ direction. An important future goal in the study of initiator proteins is to understand the architecture of an. Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. They are representatives of a large group of. Coli, there is a single origin of replication on its circular chromosome. It initiates on such dna by. After binding of two enzyme molecules (or complexes) to two recognition. Web dna unwinding enzymes use the energy released by atp hydrolysis for separating the strands of a dna. When the interactions in the dimer interface for the e323 variants are reduced,. Web in the case of e. Web open in viewer. They are representatives of a large group of. Web match these enzymes involved in dna replication with their function. In dna, consecutive nucleotides are linked via ______ bonds, which are made between the 5'. In biology there is a central dogma whereby dna is transcribed to rna, and rna is translated into protein. Substrates employed were dna, rna. Leading and lagging strands and okazaki fragments. In eukaryotic cells, there may be many thousands of origins of. It initiates on such dna by. Roles of dna polymerases and other replication enzymes. Web answer (1 of 6): Web other articles where atp synthase is discussed: Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental dna while also hydrolyzing atp. Web a similar mechanism has recently been proposed for dna unwinding by the hcv ns3 helicase in which a conformational change in the translocating enzyme occurs once. Web upon atp hydrolysis, each bapif1 molecule can unwind dna in a 5ʹ to 3ʹ direction. We have a restriction enzyme d n a lie. Coli, there is a single origin of replication on its circular chromosome. They are representatives of a large group of. Web model for the activation of restriction enzymes that require atp (or gtp) for dna cleavage. Leading and lagging strands and okazaki fragments. Web dna unwinding enzymes use the energy released by atp hydrolysis for separating the strands of a dna. Web in the case of e. Roles of dna polymerases and other replication enzymes. In biology there is a central dogma whereby dna is transcribed to rna, and rna is translated into protein. In eukaryotic cells, there may be many thousands of origins of. An important future goal in the study of initiator proteins is to understand the architecture of an initiator oligomer bound to a complete replication origin. Coli, there is a single origin of replication on its circular chromosome. It initiates on such dna by. Prior to the replication fork, helicases are enzymes that catalyse the unwinding of parental dna while also hydrolyzing atp. Web a similar mechanism has recently been proposed for dna unwinding by the hcv ns3 helicase in which a conformational change in the translocating enzyme occurs once. Web upon atp hydrolysis, each bapif1 molecule can unwind dna in a 5ʹ to 3ʹ direction. When the interactions in the dimer interface for the e323 variants are reduced,. Adenosine triphosphate:.is produced by the enzyme atp synthase, which converts adp and phosphate to atp.Which enzyme is responsible for relieving the torsional stress caused
DNA structure and replication Page 3 of 3 QCE Biology Revision
ATP molecule in an enzyme's active site, illustration Stock Image
DNA unwinding by RecBCD enzyme and structure of the enzyme bound to a
Untitled Document [www.ucl.ac.uk]
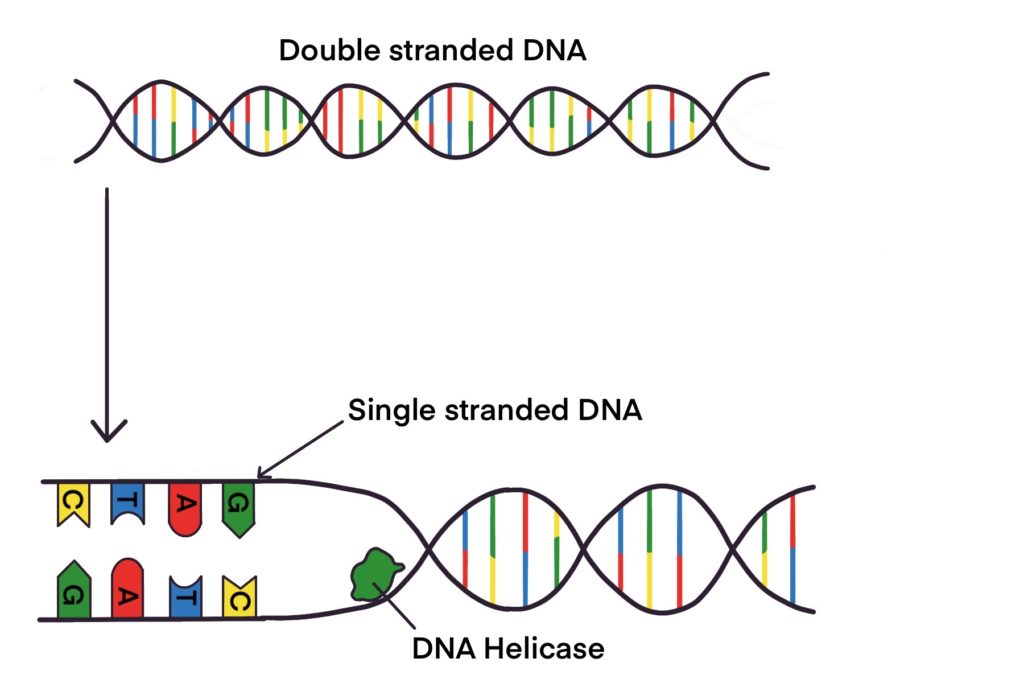
What enzyme is used to unwind DNA? Socratic
Molecular Biology 17
Enzymes of DNA replication innocent tutor
What Is Helicase? And How It unwinds DNA? Education
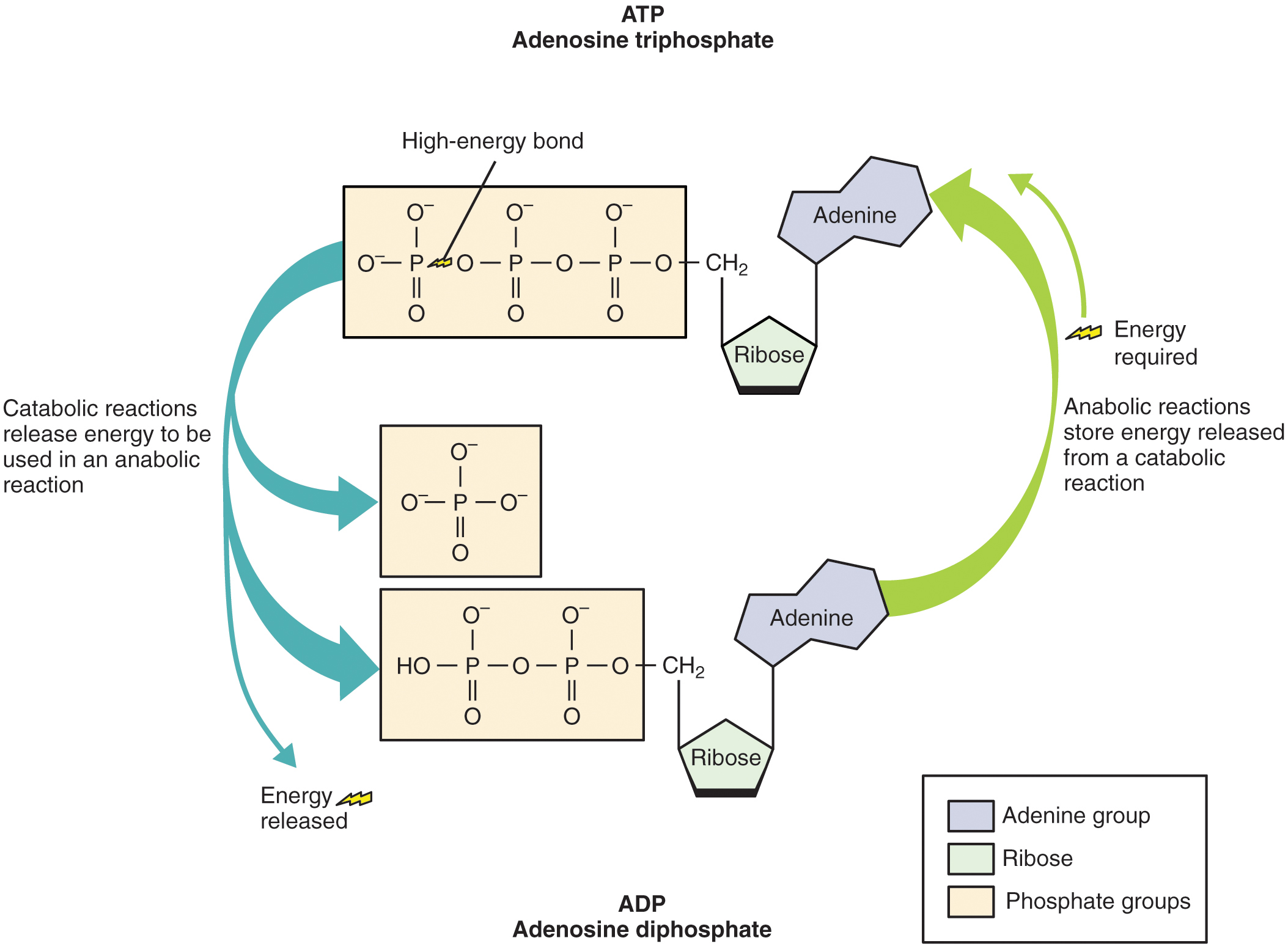
24.1 Overview of Metabolic Reactions Medicine LibreTexts
The Enzyme Used To Unwind The Dna Strands During Dna Replication, So That Each Strand Can Serve As A Template For Making A New Strand Is Dna.
After Binding Of Two Enzyme Molecules (Or Complexes) To Two Recognition.
Web Answer (1 Of 6):
Web Open In Viewer.
Related Post:




![Untitled Document [www.ucl.ac.uk]](https://www.ucl.ac.uk/~ucbcdab/oxphos/images/ATP.png)